Introduction
Biochemically, recombination of virus and bacteria is a process of creating new genomic molecules by combining or substituting fragments of nucleic acids. Genetically, recombination could be defined as the physical exchange of fragments between the parental genetic material. The results of recombination are progeny genomes that contain genetic information in non-parental combinations.
Recombination was recognized as an important factor producing genetic diversity on which natural selection can operate. Recombination events can occur in both RNA and DNA viruses. Since the molecular events behind DNA and RNA recombination differ in many respects.
Recombination in DNA viruses
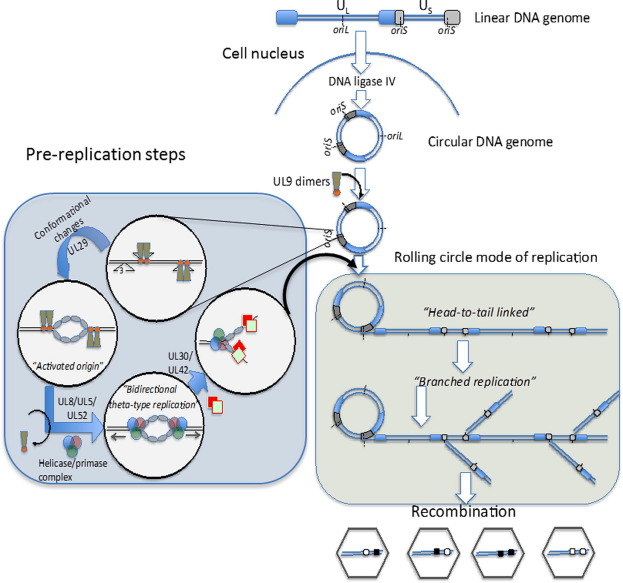
Recombination in many DNA viruses is believed to be achieved by cellular enzymatic activities. There are two general types of genetic DNA recombination in the cell: homologous recombination (general recombination) and non-homologous recombination. Non-homologous or site-specific recombination occurs relatively rarely and requires special proteins that recognize specific DNA sequences to promote recombination. Homologous recombination occurs between two DNA sequences that are the same or very similar in the region of junctions.
Homologous recombination probably occurs in all DNA-based organisms, and it happens much more often than non-homologous recombination. There is much information about the biochemical pathways responsible for crossovers in DNA. In addition to sequence identity, the requirements of general recombination include complementary base pairing between double-stranded DNA molecules, recombination enzymes, and the formation of heteroduplexes within complementary base pairing regions between the two DNA molecules recombinant.
Studies of the enzymology of DNA recombination in bacteria (and in particular of the Escherichia coli RecA, RecBCD proteins) have given rise to a large body of literature, including many general reviews. Related recombination activities have been found and studied in eukaryotic sources, including yeast, insect, mammalian, and plant cells.
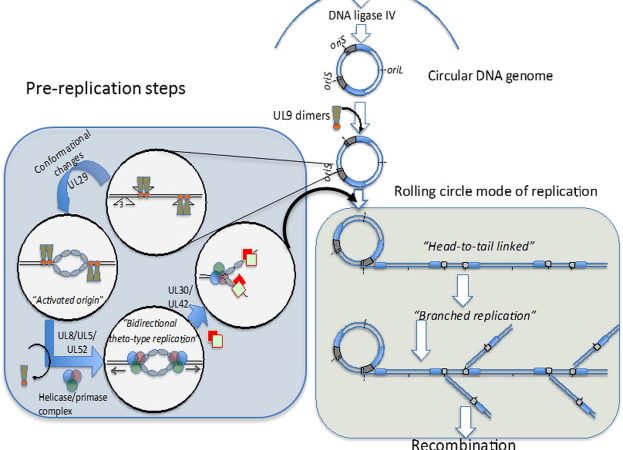
Some DNA viruses encode their own proteins that function during recombination processes. Indeed, some DNA viruses serve as model systems for the study of recombination. For example, certain bacteriophages encode recombination pathways to avoid dependency on host systems. Such recombination can be used to repair damaged phage DNA and to exchange DNA between related phages to increase their diversity.
High-frequency illegitimate recombination was observed at the origin of replication of bacteriophage M13 in host E. coli. The crossovers occurred at the nucleotide adjacent to the nick in the origin of replication, joining a nucleotide elsewhere in the genome. This implicated an illegitimate recombination break-and-rejoin mechanism, which operates in E. coli.
Recombination in RNA viruses
RNA viruses use RNA as their genetic material. The potential for variation of the RNA genome is very large due to a high rate of mutation (during copying by RNA-dependent RNA polymerase) and recombination. Classical population biology terms do not describe RNA viruses. Instead, a quasispecies term was proposed to reflect the nature of RNA virus populations. Genetic recombination processes in positive-stranded RNA viruses probably occur at the RNA level, since these viruses most likely do not go through DNA steps in their replication cycles.
RNA recombination processes are generally classified as either homologous or non-homologous. In 1992, Lai postulated three classes of RNA recombination: homologous, aberrant homologous, and nonhomologous. Homologous recombination occurs between two related RNA molecules at corresponding sites, although homologous RNA recombination can also occur within a common region shared by otherwise unrelated RNA sequences. Aberrant homologous recombination involves crossovers between related RNAs but does not occur at corresponding sites, leading to sequence insertions or deletions.
Nonhomologous recombination occurs between unrelated RNA molecules. Several authors have recently proposed slightly different definitions, based on models and mechanistic considerations. Although genetic recombination has been described in RNA viruses such as influenza virus and poliovirus, it has not been found in Newcastle disease virus. The complete nucleotide sequences of the genomic RNAs of a large number have been obtained.
Bacterial recombination mechanisms
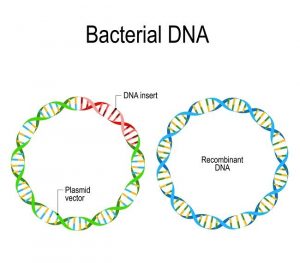
Bacterial recombination undergoes several different processes. Processes include transformation, transduction, conjugation, and homologous recombination. Homologous recombination is based on cDNA transferring genetic material. Complementary DNA sequences carry genetic material on identical homologous chromosomes. The paternal and maternal chromosomes will align so that the DNA sequences undergo the crossing over process.
Transformation involves the uptake of exogenous DNA from the surrounding environment. DNA fragments from a degraded bacterium will be transferred to the surrounding competent bacterium, resulting in an exchange of recipient DNA. Transduction is associated with virus-mediated vectors that transfer DNA material from one bacterium to another within the genome. Bacterial DNA is placed into the bacteriophage genome through bacterial transduction.
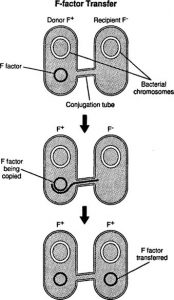
In bacterial conjugation, DNA is transferred through cell-to-cell communication. Cell-to-cell communication may involve plasmids that allow DNA transfer to another neighbouring cell. Neighboring cells take up the F plasmid (fertility plasmid: inherited material that is present on the chromosome). The recipient and donor cell come into contact during an F-plasmid transfer. The cells undergo a horizontal gene transfer in which genetic material is transferred.
Conclusions
Genetic recombination can be found in many groups of DNA and RNA viruses. Both observed natural sequence rearrangements and data obtained using experimental systems demonstrate that recombination plays an important role in providing genetic diversity during viral infections. The molecular mechanisms involved in genetic recombination depend on the class of virus. DNA viruses use homologous (general) recombination mechanisms available in host cells, although some DNA viruses encode their own recombination proteins. Furthermore, site-specific (non-homologous) recombination events were found for certain classes of DNA viruses.
For RNA viruses, both homologous and non-homologous recombination events were observed. Various types of copy choice mechanisms were proposed to explain the formation of RNA recombinants, and the role of special RNA signal sequences and viral proteins in recombination was noted. In some cases, an RNA cleavage and religation mechanism cannot be excluded and was experimentally demonstrated for bacteriophage Qβ. Further studies are required to obtain a more general picture of the genetic recombination pathways available for viruses, especially for RNA viruses.